Tutorial
How to synchronize your cloud data to Tableau extracts at scale
This is the story of some experiments that Christian Silva and I ran against the new Data Update REST API and of how this evolved into an extensible utility. This approach enables you to extract and propagate changes from any cloud database or data lake to Hyper Extracts hosted on Tableau Server or Tableau Cloud. We’ve published this utility as clouddb-extractor in the Hyper API Python Samples repository for you to use today.
Clouddb-extractor takes advantage of new functionality that was added in the 2021.2 release that allows you to run INSERT, UPDATE, and DELETE operations against published data sources. We have implemented a set of functions that coordinate processing between your Cloud Database, the client-side Hyper API, and the Tableau Server/Online REST API. This enables a number of data extract and data synchronization scenarios that were not possible before and may eliminate the need for full refresh in your environment.
This blog provides an overview of usage scenarios and how we implemented a more flexible approach to extract management. I also look at how this works under the hood and give a quick overview of how to extend this to provide a specific implementation for an additional database
Background
This project started with a real customer problem that I came across at the same time as we were beta testing the new Data Update API. They were experiencing very long running extracts from several large Google BigQuery datasets (150G+, 350M records per table). Alex Eskinasy has published a good piece on accelerating BigQuery extracts using the Storage Read API but the sheer volume and refresh frequency meant that an incremental refresh strategy was really going to be the only way to achieve the objectives. This gave us the perfect test case for this new functionality. Having demonstrated that this approach works with Google BigQuery, Christian and I then added support for AWS Redshift, PostgreSQL and MySQL to prove that this is extensible.
But wait: shouldn’t I just connect Live?
When connecting to a cloud data warehouse or data lake then Live Connection should always be your starting point as you will want to leverage the cloud platform for elastic scalability. But Tableau’s hybrid data architecture makes it simple to switch between Live and Extract without rebuilding workbooks from the ground up - so you can switch to Extract if required for heavily used data sets, for example:
- To manage costs - some cloud data warehouses charge per query.
- To provide additional concurrency or acceleration - some cloud data lakes are designed for high throughput not massive numbers of concurrent users and so you may want to “explore on Live Connections” but “operationalize on Extract”.
The need for more flexible incremental extracts
Today Tableau makes it straight-forward to take incremental extracts of extremely large cloud data sets. You can even do this entirely from the web from both Prep and Workbook Authoring. However, there are some key limitations about how changes are tracked that may have forced you to use full extract refreshes in the past. For example:
- Incremental extracts required the source table to be INSERT only. If you needed to go back and UPDATE or DELETE a row in the source table that has already been extracted there was no way of propagating this change via incremental refresh. (Changes were typically identified based on the last processed value for a date field as shown in the two screenshots below.)
Incremental extracts didn’t support rolling window data sets (for example, where you only want to retain the last X months of data and need to go back and DELETE old data after each extract refresh).
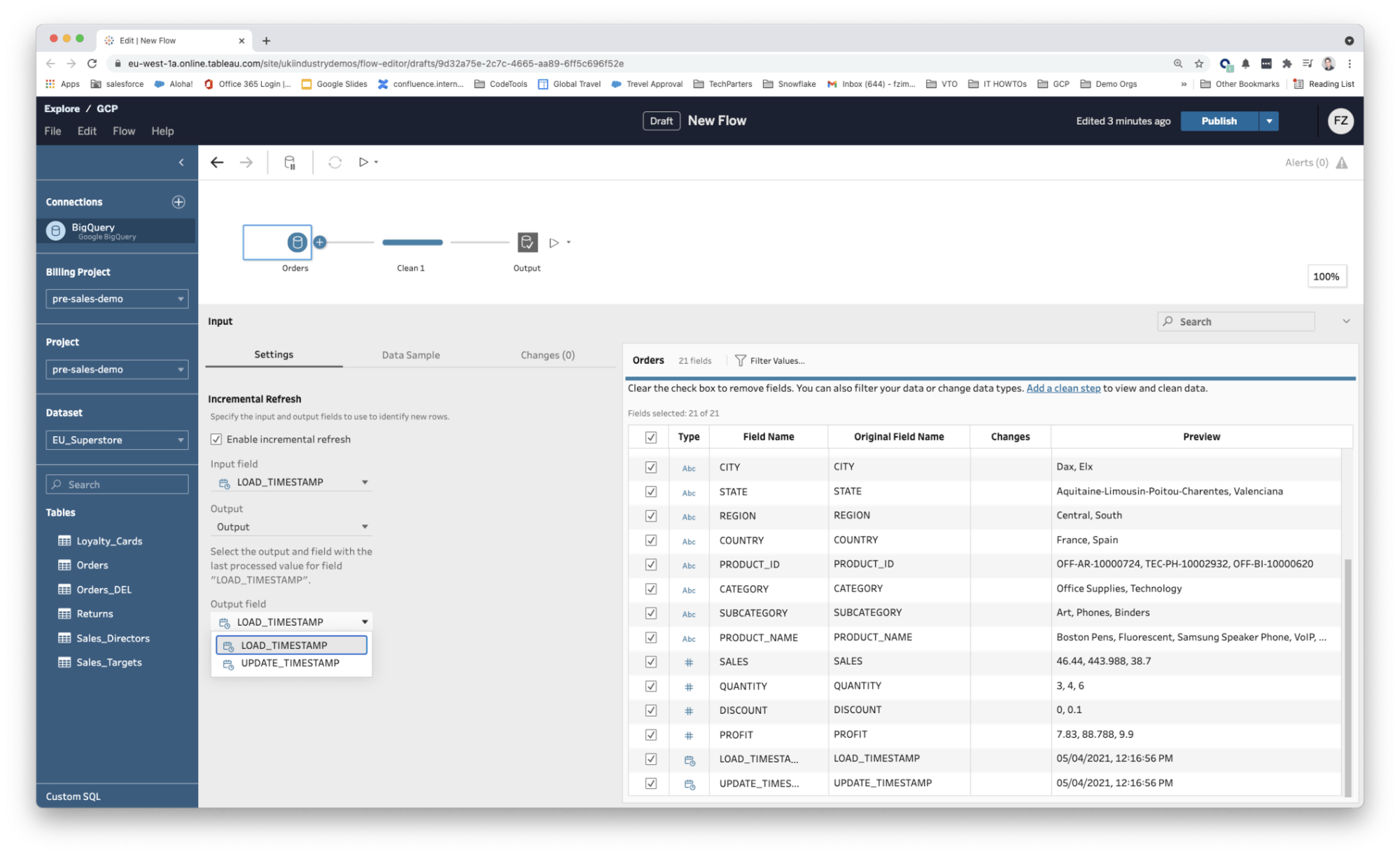
Fig 1: Managing incremental extracts using a prep flow
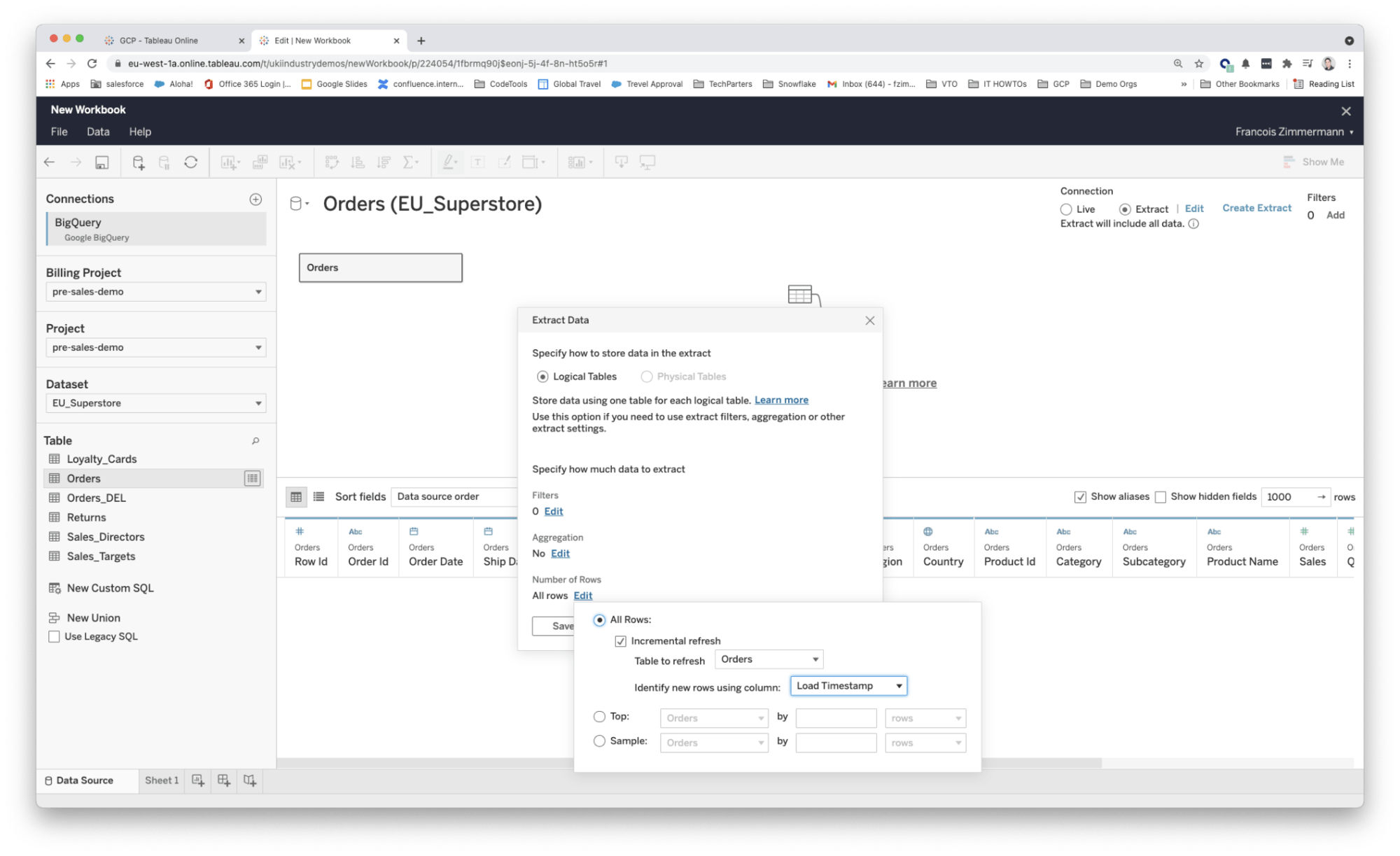
Fig 2: Managing incremental extracts from web authoring
It may be possible to overcome some of these INSERT only limitations by amending the source data model or upstream data pipeline, for example:
- Use a generated field to store [Load Date] in the source table rather than basing incremental processing on a real field like [Order Date] which may be loaded out of order if a store reports late.
- If a store needs to post an adjustment then record this in an additional row rather than updating the original.
If you’ve looked at these workarounds and determined that they still don’t meet your requirements then keep reading because you need the Data Update API (and that is why we built this utility).
Flexible incremental extract processing
Changed data tracking
Before we look at how the Data Update API will refresh our target extract we are first going to have to determine how to identify changes in the source table. We are going to need three changesets:
- INSERTED - New rows that have been added to the source table
- UPDATED - Rows that have been updated since the last successful refresh
- DELETED - Primary key columns identifying rows that have been deleted from the source table and need to be purged from the extract
Changed Data Tracking can be implemented in a number of ways, for example:
- Write a separate log of INSERTED, UPDATED, and DELETED rows during data processing.
- If you only need to cope with INSERT and UPDATE processing you could create a second generated field to store [Update Date] for incremental processing, for example: SELECT * INTO UpdatedRows FROM Orders WHERE [Update Date]>[Load Date] AND [Update Date]>&last_successful_extract
Typical workflow
After you’ve defined your strategy to track changes you need to handle workflow across a number of different layers as shown in the following image. The clouddb-extractor utility handles this processing so that you don’t have to but here is a summary of the flow from cloud database to the extract processing client to Tableau Server / Online:
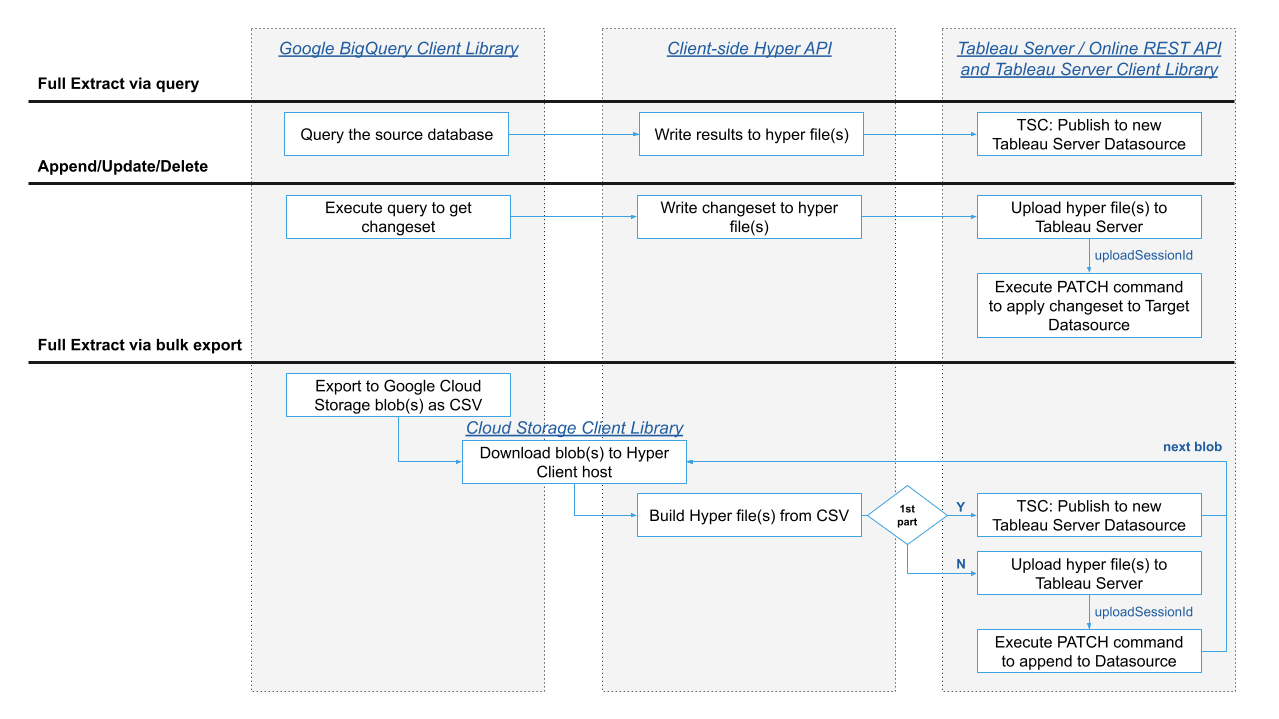
Fig 3: High-level extract processing workflow example
Extract processing methods
The clouddb-extractor utility defines a standard Extractor Interface (BaseExtractor), which is extended by specific implementations to support specific cloud databases (such as BigQueryExtractor). For most use cases you will probably only ever call the following methods to manage extract processing to published data sources on Tableau Server or Tableau Cloud :
| Method | CLI Command | Description |
|---|---|---|
| load_sample |
load_sample |
Extract a sample subset of rows from the source table to a new published datasource (Used during testing) |
| export_load |
export_load |
Full extract of source table to a new published datasource (Used for initial load) |
| append_to_datasource |
append |
Append rows from a query or table to an existing published datasource |
| update_datasource |
update |
Update an existing published datasource with the changeset from a query or table |
| delete_from_datasource |
delete |
Delete rows from a published datasource that match a condition and/or that match the primary keys in the changeset from a query or table |
We’ve included a command-line utility extractor_cli which is the simplest way to get started and test these methods out in your environment:
$ python extractor_cli.py --help
For more flexibility during automation we suggest you import the required extractor implementation and call this directly, for example:
Refer to the docstrings in the extractor class for a full list of constructor options and methods.
Using the Command Line Utility
The command line utility (extractor_cli) provides a simple way to execute these extract processing methods. The utility expects one positional argument that specifies the extract processing method to be invoked from the list above. The following named arguments are used to control behaviour:
| Named Argument | Description |
|---|---|
| --extractor [] |
Select the extractor implementation that matches your database (current options are: bigquery, redshift, mysql or postgres) |
| --source_table_id [] |
Fully qualified table identifier from source database |
| --sample_rows [] |
Defines the number of rows to use with LIMIT when command is load_sample |
| --sql [] |
The query string used to generate the changeset when command is append, update or merge |
| --sqlfile [] |
File containing the query string used to generate the changeset when command is append, update or delete. |
| --match_columns [] [] |
Define matching source and target key columns to use when command is update or delete. Specify one or more column pairs in the format: --match_columns [source_col] [target_col] |
| -match_conditions_json [] |
Json file defining conditions for matching rows when command is update or delete |
| --tableau_hostname [] |
Connection string to Tableau Server or Online |
| --tableau_site_id [] |
Target site id, use “” if connecting to default |
| --tableau_project [] |
Target project name |
| --tableau_datasource [] |
Target datasource name |
| --tableau_username [] |
Tableau username (prompts for password), required unless Personal access token is used |
| --tableau_token_name [] |
Personal access token name |
| --tableau_token_secretfile [] |
File containing personal access token secret |
CLI configuration
Before using the command line utility you will need to define your environment defaults and database connector arguments in the config.yml file as follows:
#Configuration file for clouddb_extractor utilities default_extractor : "bigquery" sample_rows : 1000 # # of rows to extract when using load_sample to preview a table tableau_env: # Tableau environment configuration defaults server_address : "https://eu-west-1a.online.tableau.com/" site_id : "testsiteid" project : "HyperAPITests" bigquery: #BigQueryExtractor configuration defaults staging_bucket : "HyperAPITestStaging" # Bucket for extract staging mysql: #Mysql configuration defaults connection: host : "mysql.test" database : "dev" port : 3306 username : "test" password : "password" raise_on_warnings : True postgres: #PostgreSQL configuration defaults connection: dbname : "dev" username : "test" password : "password" host : "postgres.test" port : 5432 redshift: #Redshift configuration defaults connection: host : 'redshift-cluster-1.XXX.eu-west-1.redshift.amazonaws.com' database : 'dev' user : 'test' password : 'password'
Command line arguments override defaults specified in the config.yml file.
Sample usage
The following examples show command line utility usage and expected output:
Load Sample
Reads the first 1000 rows from the source BigQuery table and writes to a new published Tableau datasource:
$ source env/bin/activate $ cd hyper-api-samples/Community-Supported/clouddb-extractor/ $ python3 extractor_cli.py load_sample --tableau_token_name hyperapitest --tableau_token_secretfile ~/TOL.token --source_table_id pre-sales-demo.EU_Superstore.Orders --tableau_project HyperAPITests --tableau_datasource blog_sample
Export Load
Bulk exports the entire contents of the source BigQuery table and writes to a new published Tableau datasource:
$ python3 extractor_cli.py export_load --tableau_token_name hyperapitest --tableau_token_secretfile ~/TOL.token --source_table_id pre-sales-demo.EU_Superstore.Orders --tableau_project HyperAPITests --tableau_datasource blog_sample
Update a published datasource
Executes a query against the source database to determine a changeset and then applies this to the published datasource.
updated_rows.sql:
SELECT * FROM `pre-sales-demo.EU_Superstore.Orders` WHERE LOAD_TIMESTAMP<UPDATE_TIMESTAMP
Apply update:
$ python3 extractor_cli.py update --tableau_token_name hyperapitest --tableau_token_secretfile ~/TOL.token --sqlfile updated_rows.sql --tableau_project HyperAPITests --tableau_datasource blog_sample --match_columns ROW_ID ROW_IDDelete records from a published datasource
Executes a query against the source database to determine a changeset and then deletes matching records from the published datasource. The changeset contains only the matching primary key columns that are used to identify rows to delete.
deleted_rows.sql:
SELECT * FROM `pre-sales-demo.EU_Superstore.Orders_DEL`
Delete matching rows:
$ python3 extractor_cli.py delete --tableau_token_name hyperapitest --tableau_token_secretfile ~/TOL.token --sqlfile deleted_rows.sql --tableau_project HyperAPITests --tableau_datasource blog_sample --match_columns ROW_ID ROW_ID
Delete records from a published datasource that are older than a specified date
In this example no changeset is provided - records to be deleted are identified using the conditions specified in delete_conditions.json
{ "op": "lt", "target-col": "ORDER_DATE", "const": {"type": "datetime", "v": "2018-02-01T00:00:00Z"} }
Conditional delete:
$ python3 extractor_cli.py delete --tableau_token_name hyperapitest --tableau_token_secretfile ~/TOL.token --tableau_project HyperAPITests --tableau_datasource blog_sample --match_conditions_json=delete_conditions.json
Clouddb-extractor implementation overview
We defined an abstract base class, BaseExtractor, which implements:
- A basic set of query and table extract functions, assuming the source connector implements the Python DBAPIv2 standard
- A set of functions to manage extract writes to Hyper files and application of changesets to target published Tableau data sources
The simplest way to add support for a specific database is to create a class that extends BaseExtractor and implements the following abstract methods:
| Method | Description |
|---|---|
| hyper_sql_type(source_column) |
Finds the corresponding Hyper column type for the specified source column |
| hyper_table_definition(source_table, hyper_table_name) |
Runs introspection against the named source table and returns a Hyper table definition |
| source_database_cursor() |
Handles authentication and other implementation specific dependencies or optimizations (e.g. arraysize) and returns a DBAPI Cursor to the source database. |
We made a decision not to implement generic handlers for these three methods. This is because there is not a great deal of standardization for how specific database types are reported by the Python Database API specification for the Cursor.description attribute. Also, authentication and connection options vary significantly by database, unless you are willing to be limited to basic username and password authentication.
How to Implement a new bare bones extractor
Let’s look at the PostgreSQL Extractor implementation as an example of how to implement the minimal methods required to get the utilities up and running for a new database. The source code for this bare bones implementation is here.
By implementing the above abstract methods you are just answering two questions:
1. How do I determine the structure of a source table or query and map this to a corresponding hyper table schema?
To answer this question you need to implement two methods that run introspection against the source database table or query output:
- hyper_sql_type returns the Hyper API SqlType that a source column should be mapped to.
- hyper_table_definition reads a table descriptor from the source_database and returns a Hyper API TableDefinition object.
Here is an example of how we have implemented these methods for PostgreSQL:
def hyper_sql_type(self, source_column: Any) -> SqlType: """ Finds the corresponding Hyper column type for source_column source_column (obj): Instance of DBAPI Column description tuple Returns a tableauhyperapi.SqlType Object """ type_lookup = { 16: SqlType.bool(), 17: SqlType.bytes(), 1082: SqlType.date(), 1114: SqlType.timestamp(), 20: SqlType.big_int(), 21: SqlType.int(), 1700: SqlType.numeric(18, 9), 701: SqlType.double(), 1043: SqlType.text(), 1182: SqlType.time(), 1184: SqlType.timestamp_tz(), } source_column_type = source_column.type_code return_sql_type = type_lookup.get(source_column_type) if return_sql_type is None: error_message = "No Hyper SqlType defined for Postgres source type: {}".format(source_column_type) logger.error(error_message) raise HyperSQLTypeMappingError(error_message) logger.debug("Translated source column type {} to Hyper SqlType {}".format(source_column_type, return_sql_type)) return return_sql_type def hyper_table_definition(self, source_table: Any, hyper_table_name: str = "Extract") -> TableDefinition: """ Build a hyper table definition from source_schema source_table (obj): Source table (Instance of DBAPI Cursor Description) hyper_table_name (string): Name of the target Hyper table, default="Extract" Returns a tableauhyperapi.TableDefinition Object """ # logger.debug( # "Building Hyper TableDefinition for table {}".format(source_table.dtypes) # ) target_cols = [] for source_column in source_table: this_name = source_column.name this_type = self.hyper_sql_type(source_column) this_col = TableDefinition.Column(name=this_name, type=this_type) target_cols.append(this_col) logger.info("..Column {} - Type {}".format(this_name, this_type)) # create the target schema for our Hyper File target_schema = TableDefinition(table_name=TableName("Extract", hyper_table_name), columns=target_cols) return target_schema
2. How do I connect to the source database and obtain a cursor to execute queries?
To answer this question you need to implement the source_database_cursor method which defines authentication options for your source database, handles connection errors and configures the DBAPI cursor. For example we configure PostgreSQL to provide a server side cursor rather than the default client side cursor in order to limit memory use for very large data sets:
def source_database_cursor(self) -> Any: """ Returns a DBAPI Cursor to the source database """ if self._source_database_connection is None: db_connection_args = self.source_database_config.get("connection") self._source_database_connection = psycopg2.connect(**db_connection_args) # Use Server Side Cursor: # If the dataset is too large to be practically handled on the client side, # it is possible to create a server side cursor. Using this kind of cursor # it is possible to transfer to the client only a controlled amount of data, # so that a large dataset can be examined without keeping it entirely in memory. # Psycopg wraps the database server side cursor in named cursors. # A named cursor is created using the cursor() method specifying the name parameter. named_server_side_cursor = self._source_database_connection.cursor(name=uuid.uuid4().hex) return named_server_side_cursor
Defining technology-specific performance optimizations
The default behavior of the utility is to execute all extracts via the DBAPIv2 Cursor.fetchmany() method. However, your cloud database vendor might provide more performant native client libraries to transfer data. For example, BigQuery provides mechanisms to estimate query size before execution (useful for cost control) and also enables you to export data via Cloud Storage (much faster for large tables). Here is an example of how we override the query_to_hyper_files method in order to implement database specific optimization (full source code here):
def _estimate_query_bytes(self, sql_query: str) -> int: """ Dry run to estimate query result size """ # Dry run to estimate result size job_config = bigquery.QueryJobConfig(dry_run=True, use_query_cache=False) dryrun_query_job = bq_client.query(sql_query, job_config=job_config) dryrun_bytes_estimate = dryrun_query_job.total_bytes_processed logger.info("This query will process {} bytes.".format(dryrun_bytes_estimate)) if dryrun_bytes_estimate > MAX_QUERY_SIZE: raise QuerySizeLimitError("This query will return more than {MAX_QUERY_SIZE} bytes") return dryrun_bytes_estimate def query_to_hyper_files( self, sql_query: Optional[str] = None, source_table: Optional[str] = None, hyper_table_name: str = "Extract", ) -> Generator[Path, None, None]: """ Executes sql_query or exports rows from source_table and writes output to one or more hyper files. Returns Iterable of Paths to hyper files sql_query (string): SQL to pass to the source database source_table (string): Source table ref ("project ID.dataset ID.table ID") hyper_table_name (string): Name of the target Hyper table, default=Extract NOTES: - Specify either sql_query OR source_table, error if both specified """ extract_prefix = "" extract_destination_uri = "" use_extract = False target_table_def = None if not (bool(sql_query) ^ bool(source_table)): raise Exception("Must specify either sql_query OR source_table") if sql_query: assert source_table is None self._estimate_query_bytes(sql_query) if USE_DBAPI: logging.info("Executing query using bigquery.dbapi.Cursor...") for path_to_database in super().query_to_hyper_files( sql_query=sql_query, hyper_table_name=hyper_table_name, ): yield path_to_database return else: logging.info("Executing query using bigquery.table.RowIterator...") query_job = bq_client.query(sql_query) # Determine table structure target_table_def = self.hyper_table_definition(source_table=bq_client.get_table(query_job.destination), hyper_table_name=hyper_table_name) query_result_iter = bq_client.list_rows(query_job.destination) path_to_database = self.query_result_to_hyper_file( target_table_def=target_table_def, query_result_iter=query_result_iter, ) yield path_to_database return else: logging.info("Exporting Table:{}...".format(source_table)) use_extract = True extract_prefix = "staging/{}_{}".format(source_table, uuid.uuid4().hex) extract_destination_uri = "gs://{}/{}-*.csv.gz".format(self.staging_bucket, extract_prefix) source_table_ref = bq_client.get_table(source_table) target_table_def = self.hyper_table_definition(source_table_ref, hyper_table_name) extract_job_config = bigquery.ExtractJobConfig(compression="GZIP", destination_format="CSV", print_header=False) extract_job = bq_client.extract_table(source_table, extract_destination_uri, job_config=extract_job_config) extract_job.result() # Waits for job to complete. logger.info("Exported {} to {}".format(source_table, extract_destination_uri)) if use_extract: bucket = storage_client.bucket(self.staging_bucket) pending_blobs = 0 batch_csv_filename = "" for this_blob in bucket.list_blobs(prefix=extract_prefix): logger.info("Downloading blob:{} ...".format(this_blob)) # # TODO: better error checking here temp_csv_filename = tempfile_name(prefix="temp", suffix=".csv") temp_gzip_filename = "{}.gz".format(temp_csv_filename) this_blob.download_to_filename(temp_gzip_filename) logger.info("Unzipping {} ...".format(temp_gzip_filename)) # Performance optimization: Concat smaller CSVs into a larger single hyper file if pending_blobs == 0: # New batch batch_csv_filename = temp_csv_filename subprocess.run( f"gunzip -c {temp_gzip_filename} > {batch_csv_filename}", shell=True, check=True, ) os.remove(Path(temp_gzip_filename)) else: # Append to existing batch subprocess.run( f"gunzip -c {temp_gzip_filename} >> {batch_csv_filename}", shell=True, check=True, ) os.remove(Path(temp_gzip_filename)) pending_blobs += 1 if pending_blobs == BLOBS_PER_HYPER_FILE: path_to_database = self.csv_to_hyper_file( path_to_csv=batch_csv_filename, target_table_def=target_table_def, ) pending_blobs = 0 os.remove(Path(batch_csv_filename)) yield path_to_database if pending_blobs: path_to_database = self.csv_to_hyper_file(path_to_csv=batch_csv_filename, target_table_def=target_table_def) os.remove(Path(batch_csv_filename)) yield path_to_database
Next steps
This is community maintained code and we welcome any input. If you would like to provide feedback or suggest enhancements then please raise an issue in the GitHub repository here. Be sure to include clouddb-extractor in the title. In particular, please let us know if you need an implementation for a specific cloud database.
To learn more about the Data Update REST API, see the documentation here. You can also see more content by signing up for our Developer Program.
Senaste uppdatering: 13 September, 2021
